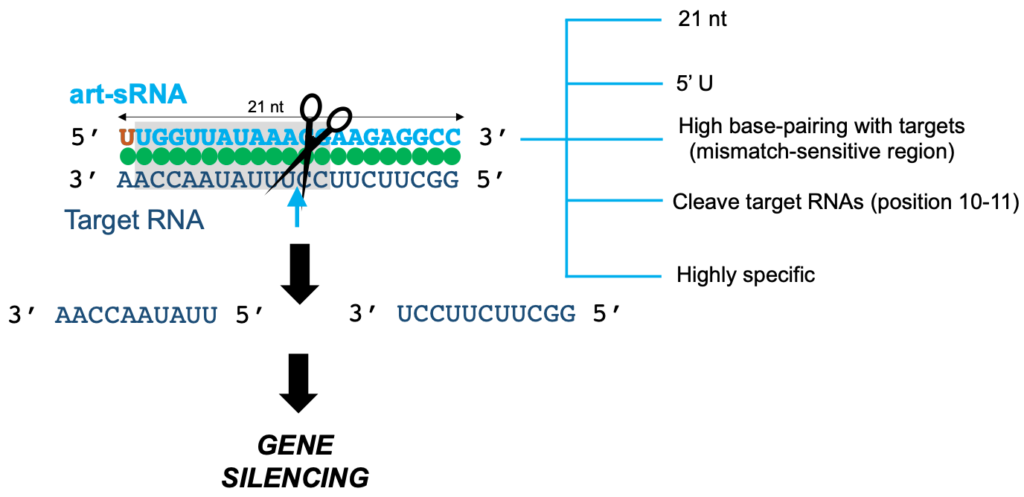
In most eukaryotes, gene expression can be suppressed through the sequence-specific degradation of target RNA by complementary small RNAs (sRNAs), a process termed RNA interference (RNAi). Recent RNAi tools are based on sRNAs that are designed to silence specific plant genes and pathogens. Such sRNAs are named artificial sRNAs (art-sRNAs) which are typically 21-nt long and computationally designed to silence target RNA with high efficacy and specificity.
Our lab seeks to develop next-generation art-sRNAi tools for controlling gene expression and inducing antiviral resistance in plants.
We have the following specific goals:
(i) Optimizing art-sRNAi efficacy, fine-tunability and systemicity for absolute control of plant gene expression.
(ii) Developing efficient GMO-free art-sRNAi through the cost-efficient production and topical delivery of art-sRNA precursors to plants, and to use viral vectors or by editing endogenous sRNA precursors to express art-sRNAs.
(iii) Applying these optimized methodologies to improve real crops.
Some of our latest scientific achievements:
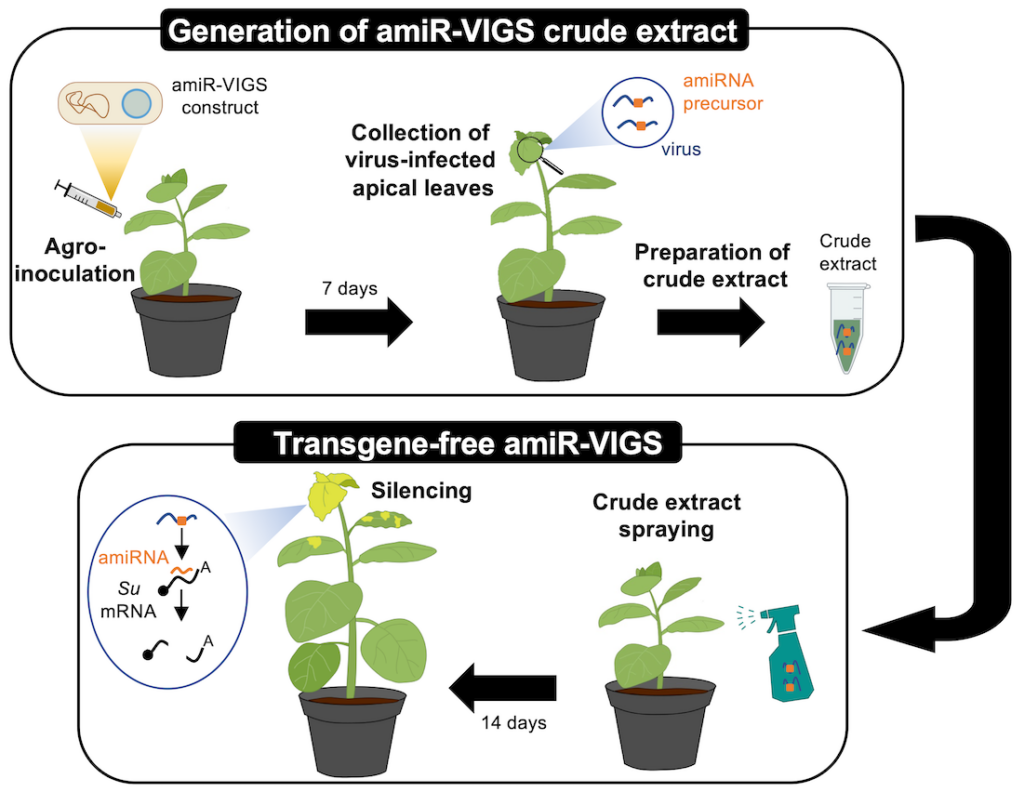
Transgene-free, virus-based gene silencing in plants by artificial microRNAs. We show that gene silencing efficacy of AtMIR390a-derived amiRNAs is maintained in when expressed from a minimal, chimeric precursor of only 89 nt. Remarkably, minimal but not full-length precursors produce authentic amiRNAs and induce widespread gene silencing in Nicotiana benthamiana when expressed from an RNA virus such as Potato Virus X, which can be applied into leaves by spraying infectious crude extracts. This methodology allows the application of amiRNAs to plants in a transgene-free manner for whole-individual gene silencing .
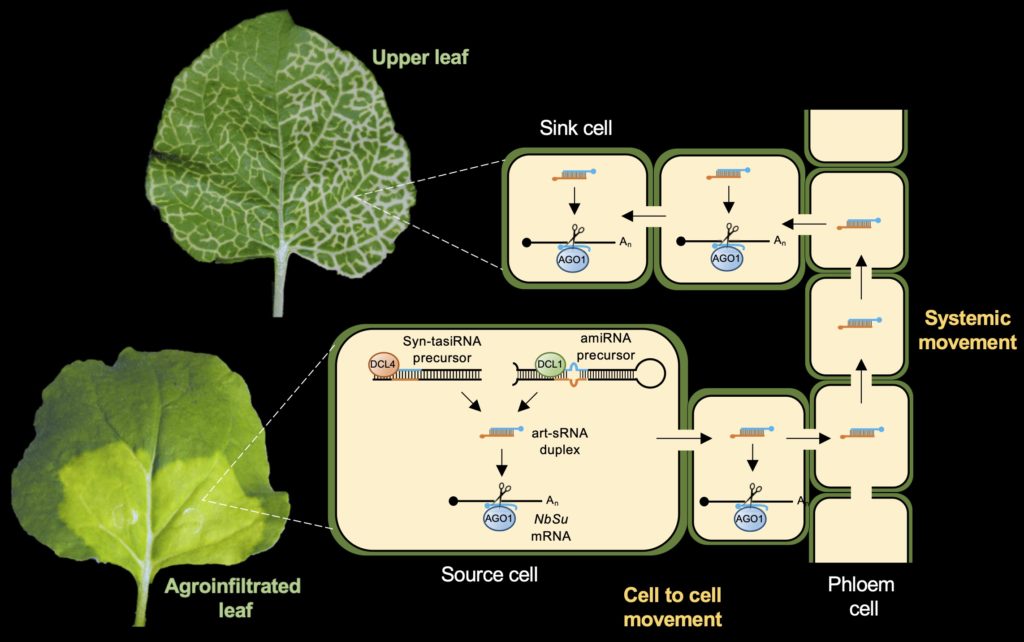
Systemic silencing of plant genes triggered by art-sRNAs. We have demonstrated that 21-nt art-sRNAs can move throughout the plant to silence plant genes in tissues different from where they are produced. This highlights the biotechnological potential of art-sRNAs, which might be applied locally for triggering whole-plant and highly specific silencing to regulate gene expression or induce resistance against pathogenic RNAs in next-generation crops.
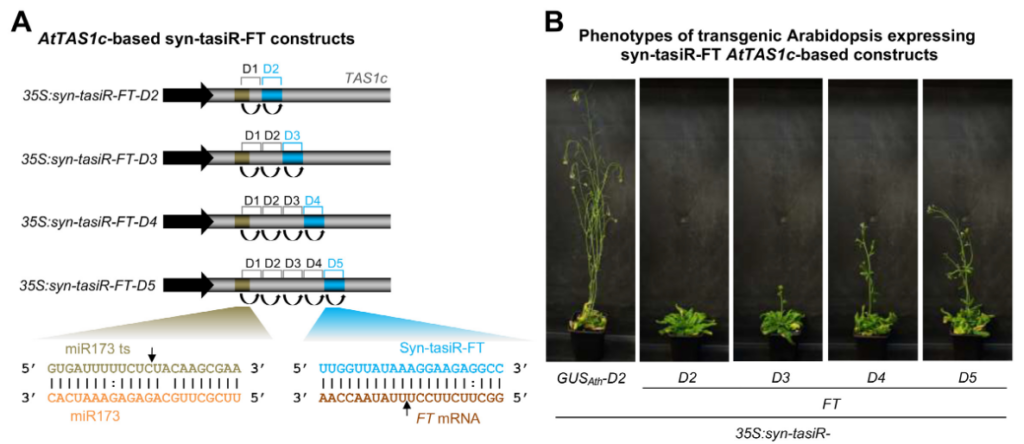
Fine-tune modulation of gene expression in plants by art-sRNAs. We developed two different strategies based on art-sRNAs for fine-tuning the degree of induced silencing in plants: i) by modifying the precursor position from which a syn-tasiRNA is expressed, and ii) by modifying the degree of base-pairing between the 3’ end of the syn-tasiRNA and the 5’ end of the target RNA. Both strategies were used to finely modulate the degree of silencing of endogenous and exogenous target genes in A. thaliana and N. benthamiana.
Development of tomato plants expressing multiple syn-tasiRNAs and highly resistant to Tomato spotted wilt virus. We showed that syn-tasiRNAs induce enhanced antiviral resistance because of the combined silencing effect of each individual syn-tasiRNA, which minimizes the possibility that the virus simultaneously mutates all different target sites to fully escape each syn-tasiRNA.
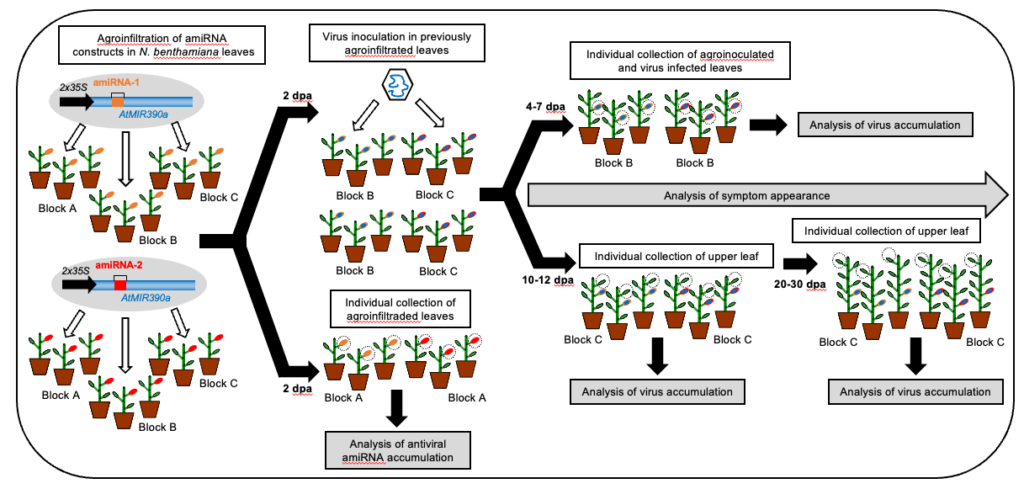
Development of a high-throughput methodology to identify highly effective art-sRNAs against plant viruses and viroids. We developed a systematic methodology for the simple and fast-forward design, generation and functional analysis of large numbers of art-sRNA constructs engineered to induce high levels of antiviral resistance in plants. Art-sRNA constructs are transiently expressed in N. benthamiana plants, which are subsequently inoculated with the virus or viroid of interest. The antiviral activity of each art-sRNA construct is assessed by monitoring viral (viroid) symptom appearance, and through molecular analysis of virus (viroid) accumulation in plant tissues. This approach is aimed to easily identify art-sRNAs with high antiviral activity that could be expressed in transgenic plants for highly durable antiviral.